Unveiling the Molecular "Superpower" of Antibiotic-Resistant Bacteria
A recent study conducted by Lund University in Sweden has shed light on the molecular mechanisms that enable a specific species of gut bacteria to thrive and become highly resistant to antibiotics when the natural intestinal flora is disrupted by antibiotic treatment. This resilient bacterium, which is naturally resistant to many antibiotics, poses significant challenges, particularly in healthcare settings. The research team discovered two molecular mechanisms that work in tandem to enhance the bacterium's resistance. The findings have the potential to aid the development of more effective medications, as stated by Vasili Hauryliuk, the senior lecturer at Lund University who spearheaded the study. [1]
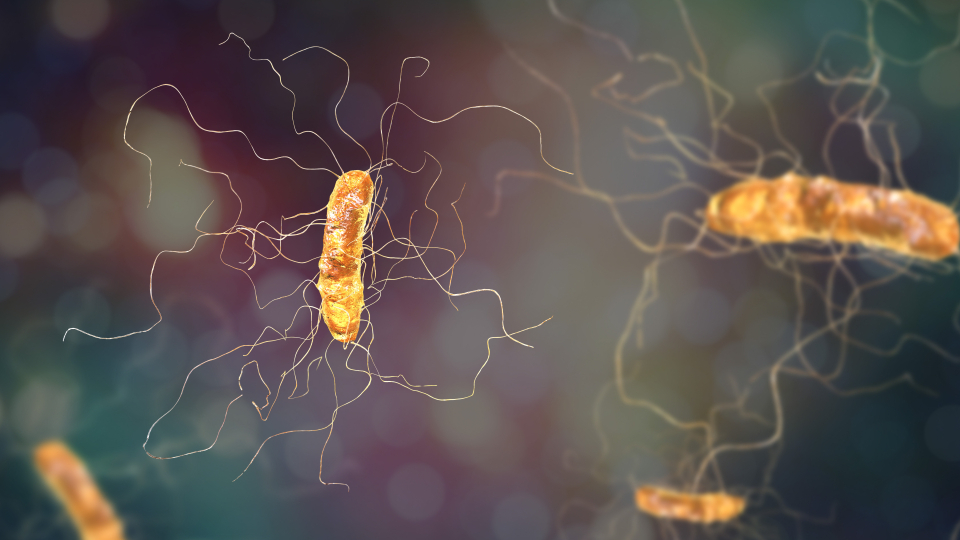
Figure 1. Clostridioides difficile bacterium
Figure 1 shows an illustration of Clostridioides difficile bacterium with peritrichous flagella. The threat posed by antibiotic-resistant bacteria is widely recognized and of significant concern. According to a report published by The Lancet last year, approximately 1.27 million individuals lost their lives in 2019 due to bacterial infections that could not be effectively treated with available medications. To effectively combat this grave threat, it is crucial to gain a comprehensive understanding of the underlying molecular mechanisms that contribute to antibiotic resistance. [1]
Antibiotic treatment disrupts the normal gut flora, allowing antibiotic-resistant bacterial pathogens to thrive. Among these pathogens, Clostridioides difficile (C. diff) is particularly problematic. Resistant to antibiotics, it can cause severe diarrhea infections. C. diff's spore-forming ability facilitates easy transmission, leading to challenges in healthcare settings, including higher mortality rates and prolonged treatment durations. [1]
“Instead of the antibiotic saving you, in this case, it promotes a secondary bacterial infection,” says Vasili Hauryliuk. [1]
“The risk of infection with C. diff is known to increase after treatment with an antibiotic called clindamycin, but the reason for this was unknown. Our research showed a novel protein conveys resistance to the class of antibiotics to which clindamycin belongs,” says Obana Nozomu, assistant professor at the University of Tsukuba and one of the researchers behind the study. [1]
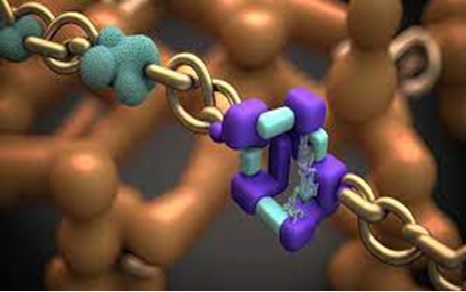
In a collaborative effort involving researchers from Sweden, Japan, the United Kingdom, USA, Estonia, and Germany, a study investigating the resistance mechanism of Clostridioides difficile (C. diff) has been conducted. The findings of this study have been published in Nucleic Acids Research. The researchers successfully identified a novel protein responsible for the resistance exhibited by C. diff. This protein operates on the ribosome, which serves as the molecular factory responsible for protein synthesis in bacteria. The ribosome is a crucial target for antibiotics, as inhibiting protein synthesis prevents bacterial growth, replication, and the ability to cause infections. The discovery of this protein sheds light on the specific mechanisms through which C. diff develops its antibiotic resistance. “This newly discovered protein kicks the antibiotic molecule out of the ribosome. We also saw that it combines with another resistance factor. The second chemically modifies the ribosome so that the antibiotic molecules to bind less tightly to it. The extra-potent resistance is the result of two mechanisms, two factors, which combine and in so doing give the bacteria its ‘superpowers’ against antibiotics,” says Gemma C. Atkinson, senior lecturer at Lund University and co-author of the article. [1]
“This newly discovered protein kicks the antibiotic molecule out of the ribosome. We also saw that it combines with another resistance factor. The second chemically modifies the ribosome so that the antibiotic molecules to bind less tightly to it. The extra-potent resistance is the result of two mechanisms, two factors, which combine and in so doing give the bacteria its ‘superpowers’ against antibiotics,” says Gemma C. Atkinson, senior lecturer at Lund University and co-author of the article. [1]
Using cryogenic electron microscopy, the researchers conducted a detailed investigation of the molecular-level resistance mechanisms employed by Clostridioides difficile (C. diff) against antibiotics. This breakthrough in understanding paves the way for the development of novel treatment strategies to combat antibiotic resistance and effectively tackle the infections caused by these bacteria. [1]
“A couple of years ago, Andrew G. Myers lab at Harvard University has developed a new generation of ribosome-binding antibiotics, known as iboxamycin. It is a very potent medicine that knocks out ‘ordinary’ C. diff bacteria. The results of this study, however, show that C. diff strains that have both resistance factors are, unfortunately, resistant to this antibiotic as well. This means that it is necessary to design antibiotic molecules that bind even tighter in order to overpower this kind of resistance. We now collaborate with the Myers group on this direction.” says Vasili Hauryliuk. [1]
This study also found that certain antibiotics that target the ribosome induce the production of the resistance factor. This may also provide clues for designing new antibiotic molecules, since resistance cannot be induced if resistance factors are not synthesized. [1]
Source: Lund University
References:
- https://www.lunduniversity.lu.se/article/revealed-molecular-superpower-antibiotic-resistant-bacteria
- https://scitechdaily.com/molecular-superpower-of-antibiotic-resistant-bacteria-revealed-in-new-research/
Cite this article:
Hana M (2023), Unveiling the Molecular "Superpower" of Antibiotic-Resistant Bacteria, AnaTechMaz,pp 193