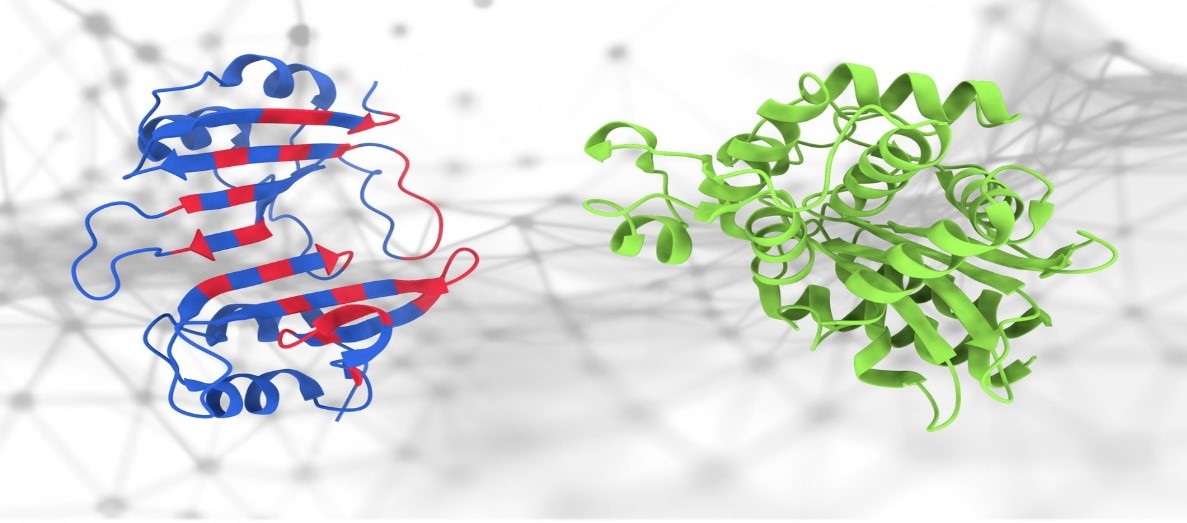
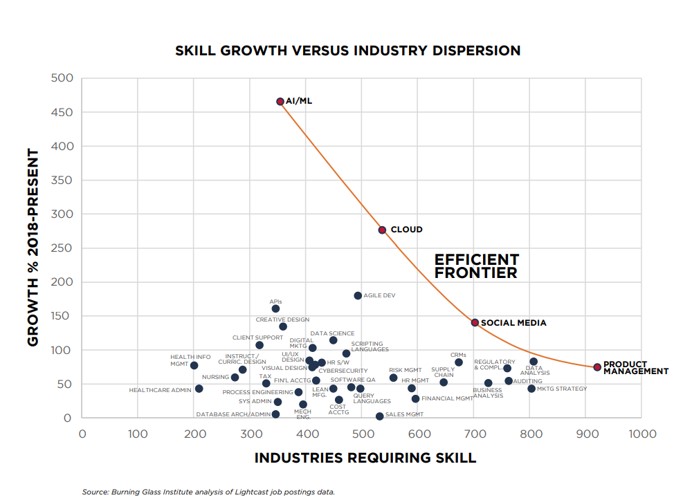
PeSTo: a new AI tool for predicting protein interactions
Scientists at EPFL have developed PeSTo, an AI model that can predict, with high confidence, the binding interfaces of proteins when they bind other proteins, nucleic acids, lipids, ions, and small molecules. The model's low computational cost enables the processing of large amounts of structural data, opening up opportunities for new biological discoveries.
In a big step forward, scientists in the group of Matteo Dal Peraro at EPFL have developed a new tool called PeSTo (short for Protein Structure Transformer) that can predict the specific regions on the surface of a protein that can interact with other proteins, but also nucleic acids, lipids, ions, and small molecules. These interfaces are crucial for the formation of supramolecular complexes and function modulation. [1]

Figure 1. PeSTo: a new AI tool for predicting protein interactions
PeSTo: a new AI tool for predicting protein interactions is shown in Figure 1. PeSTo is built on a neural network based on transformer technology. In the context of machine learning, a transformer is a type of neural network designed to process sequential data, such as natural language by using self-attention mechanisms to weigh the importance of different parts of the input sequence and make predictions. The technology was introduced by Google Brain in 2017 and is now at the core of many modern AI tools.
PeSTo applied to the human foldome
The researchers unleashed PeSTo on the “human foldome”, a growing database of predicted protein structures. They analyzed the interactions that human proteins have with other molecules, and produced detailed information about the human “interfaceome” – the sum total of all protein interacting interfaces in the human body. To do this, the researchers used the AlphaFold European Bioinformatics Institute (AF-EBI) database.
The researchers have made PeSTo available in a user-friendly web server, free of charge and prior registration. The server can take any protein structure in PDB format. The predicted interfaces can be visualized directly in the browser with additional information on the confidence of the prediction on a per-residue basis.
Publishing in Nature Communications [below], the scientists highlight numerous advantages of PeSTo over older methods, particularly that it can work with all kinds of molecules without needing to know all the details about their chemistry and physics. This makes PeSTo a more flexible, powerful and general tool for studying molecular systems and their interactions. [2]
Overall, PeSTo holds great promise as a valuable AI tool for predicting protein interactions. Its ability to accelerate our understanding of protein-protein interactions has the potential to revolutionize various fields of biology and medicine, contributing to advancements in drug discovery, personalized medicine, and synthetic biology.
However, it is important to note that PeSTo, like any predictive AI tool, has its limitations. The accuracy of predictions heavily relies on the quality and diversity of the training data. Therefore, ongoing efforts to curate high-quality and comprehensive protein interaction datasets will be crucial for improving the performance of PeSTo and similar tools.
References:
- https://actu.epfl.ch/news/pesto-a-new-ai-tool-for-predicting-protein-interac
- https://sciencesprings.wordpress.com/2023/05/10/from-the-swiss-federal-institute-of-technology-in-lausanne-epfl-ecole-polytechnique-federale-de-lausanne-ch-pesto-a-new-ai-tool-for-predicting-protein-interactions/
Cite this article:
Gokula Nandhini K (2023), PeSTo: a new AI tool for predicting protein interactions, AnaTechMaz, pp.262