Single Cell Technologies
Single cell technologies are becoming increasingly important tools in biological analysis. Complementing average measurements on bulk populations of cells, single-cell measurements provide a finer-grained picture of complex biology and unmask heterogeneity that is present in tissues. With the increasing sophistication of microfluidics, electrophysiology measurements, high-resolution imaging, deep sequencing and mass spectrometry platforms, a more detailed picture of cellular subtype, physical location in tissue, and clonal evolution is emerging. Moreover, the exquisite sensitivity of these approaches is enabling the identification of rare cells of potential functional or pathogenic consequence. This mini-focus brings together a set of articles that explores key issues in analyzing, integrating and applying single-cell data with an emphasis on challenges for the field.[1]
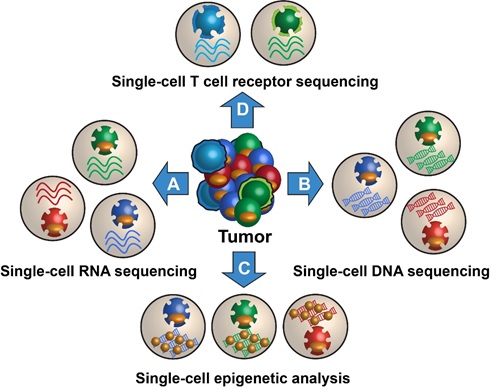
Figure 1. The Single cell Technologies in tumor
Figure 1 shows the Single-cell RNA-sequencing (scRNA-seq) has been widely adopted in recent years due to standardized protocols and automation, reliability, and standardized bioinformatic pipelines. The most widely adopted platform is the 10× Genomics solution.[2]
Brain tumor
It is generally believed that brain astrocytomas and oligodendrogliomas originate from different types of cells. However, Venteicher et al.found that IDH mutant gliomas share a common origin through single-cell sequencing technology. The difference between them is mainly the difference in genetics and the composition of the tumor microenvironment. Two of the different subtypes of tumors have universally similar stem cell programs, which can be distinguished from similar neuroglial mass spectrometry systems. Tirosh et al. [28] identified cancer stem cells and their differentiated progeny through a single cell map in human oligodendroglioma. The findings support the cancer stem cell hypothesis and confirm that cancer stem cells were the main source of growth for oligodendroglioma. These studies suggest that these cells may become new therapeutic targets.
Hematological tumors
Single-cell sequencing technologies help to find classification markers and analyze the causes of disease occurrence in hematological tumors with high heterogeneity, providing a theoretical basis for clinical diagnosis and treatment of diseases.
Ledergor et al. found at the single-cell level that there were common expressions and differences in certain genes among different myeloma plasma cell samples. New markers can be sought by these common expressions. Differences in gene expression patterns and chromosome structure among the samples may be responsible for the high heterogeneity of myeloma plasma cells. [3]
Future of Single - cell
Future progress could also include the combination of multiple omics to obtain a more comprehensive view of immune cell states. A simple, automated, high-throughput and user-friendly platform should be developed which combines mRNA sequencing with protein, genomic and epigenomic analysis to gain more cellular information compared with transcriptomic data alone. At the same time, integrating cellular imaging into studies could also provide more informative output such as cell morphology, phenotype or viability. One of the ongoing challenges in immunology is to acquire more detailed data on single immune cells in order to provide better understanding of immune cell responses to human diseases. Together, these integrated strategies will provide us with a more comprehensive view of cellular heterogeneity and immune responses to contribute significantly to future biomedical discoveries and advance human health.[4]
References:
- https://www.nature.com/collections/nylbrpjhlp/
- https://kidney360.asnjournals.org/content/2/7/1196
- https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-019-0314-y
- https://www.future-science.com/doi/10.2144/btn-2020-0047
Cite this article:
Thanusri swetha J (2021), Single Cell Technologies, AnaTechMaz, pp. 18