MIT Biologists Uncover a Novel Mechanism for Regulating RNA Splicing
MIT biologists have discovered a new regulatory layer in RNA splicing, revealing a mechanism that influences spliceosome targeting on messenger RNA. This process appears to affect about half of all human genes and is found across the animal kingdom and in plants. The findings suggest that RNA splicing is more intricate than previously thought, allowing for more sophisticated gene regulation in complex organisms like humans.

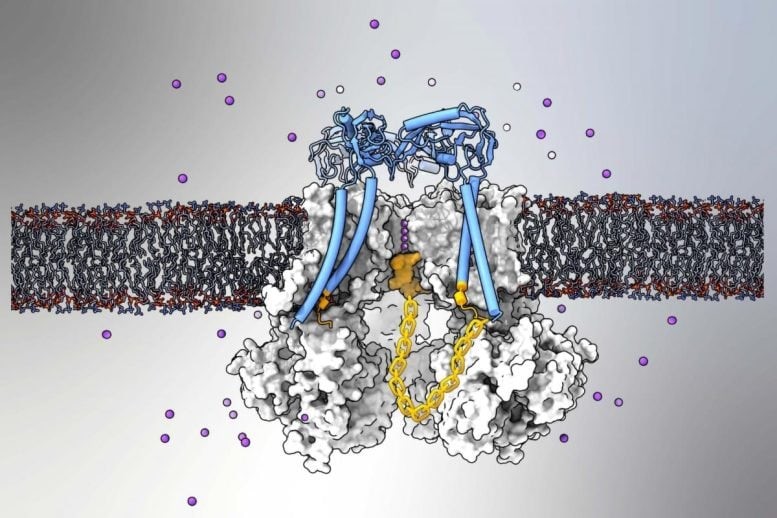
Figure 1. MIT Biologists Reveal a New Mechanism for RNA Splicing Regulation.
The research team found that this regulatory mechanism, which influences the expression of nearly half of all human genes, is widespread across the animal kingdom and present in plants. Their findings suggest that RNA splicing, a fundamental process in gene expression, is more intricate than previously understood. Figure 1 shows MIT Biologists Reveal a New Mechanism for RNA Splicing Regulation.
“Splicing in complex organisms like humans is more sophisticated than in simpler models like yeast, even though it remains a highly conserved molecular process. The human spliceosome has additional features that enable it to process specific introns more efficiently. This system may provide an advantage by allowing more intricate forms of gene regulation,” says Connor Kenny, an MIT graduate student and the study’s lead author.
Building Proteins
Discovered in the late 1970s, RNA splicing allows cells to precisely regulate the content of mRNA transcripts, which carry instructions for protein synthesis.
Each mRNA transcript consists of coding regions (exons) and noncoding regions (introns), along with specific sites that signal where splicing should occur. This mechanism enables cells to assemble the correct sequence for producing a desired protein. By allowing a single gene to generate multiple proteins, RNA splicing plays a crucial role in genetic diversity. Over evolutionary timescales, it can also alter gene and protein structure by including or excluding different exons.
Uncovering a New Layer of Splicing Regulation
The spliceosome, which assembles on introns, consists of proteins and noncoding RNAs known as small nuclear RNAs (snRNAs). During the first step of spliceosome formation, an snRNA molecule called U1 snRNA binds to the 5’ splice site at the beginning of an intron. Previously, scientists believed that the strength of this binding was the key factor determining whether an intron would be spliced out of an mRNA transcript.
In a new study, MIT researchers discovered that a family of proteins called LUC7 also plays a crucial role in splicing—specifically for a subset of introns, affecting up to 50% of human genes. While LUC7 proteins were known to associate with U1 snRNA, their exact function had remained unclear.
The study identified three distinct LUC7 proteins in human cells. Two of these proteins interact with a specific type of 5’ splice site, termed “right-handed,” while the third LUC7 protein associates with a different type, labeled “left-handed.” The researchers found that about half of human introns contain these right- or left-handed sites, while the remaining introns appear to be regulated by other mechanisms.
This discovery reveals an additional layer of regulation that enhances the efficiency of intron removal. “The paper shows that these two different 5’ splice site subclasses exist and can be regulated independently of one another,” says Connor Kenny. “Some of these core splicing processes are actually more complex than we previously appreciated, which warrants more careful examination of what we believe to be true about these highly conserved molecular processes.”
Complex Splicing Machinery
Previous research has linked mutations or deletions in one of the LUC7 proteins that bind to right-handed splice sites with blood cancers, including about 10% of acute myeloid leukemias (AMLs). In this study, MIT researchers found that AMLs with a loss of one copy of the LUC7L2 gene exhibit inefficient splicing at right-handed splice sites. These cancers also develop metabolic changes similar to those observed in earlier studies.
“Understanding how the loss of this LUC7 protein in some AMLs alters splicing could help in the design of therapies that exploit these splicing differences to treat AML,” says Christopher Burge. He notes that small-molecule drugs used for other diseases, such as spinal muscular atrophy, work by stabilizing the interaction between U1 snRNA and specific 5’ splice sites. Since LUC7 proteins influence these interactions at specific splice sites, this knowledge could enhance the specificity of such drugs.
Collaborating with a team led by Sascha Laubinger at Martin Luther University Halle-Wittenberg, the researchers also discovered that plants possess right- and left-handed 5’ splice sites regulated by Luc7 proteins. Their analysis suggests that this splicing mechanism originated in a common ancestor of plants, animals, and fungi, but was later lost in fungi after they diverged from plants and animals.
“A lot of what we know about splicing and its core components comes from early yeast genetics research,” says Connor Kenny. “What we see is that humans and plants have more complex splicing machinery, with additional components that allow for independent regulation of different introns.”
Source: MIT NEWS
Cite this article:
Priyadharshini S (2025),MIT Biologists Uncover a Novel Mechanism for Regulating RNA Splicing,AnaTechMaz,pp. 334