Profiling MicroRNA in Feces to Investigate Gut Health and Disease
The mammalian gut is a hub for digestion, immune activity, and interactions between the host and microbes. Intestinal microRNAs—tiny, non-coding molecules that control gene expression—play key roles in these processes and can be detected in feces, providing a non-invasive way to assess gut health. Microbiologist Emma Layton, during her graduate studies at The University of Manchester, aimed to explore fecal microRNAs but faced challenges due to the lack of established methods for extracting RNA from fecal samples or preparing small RNA sequencing libraries. She is now continuing this work at the University of Oxford.
To address the lack of standardized methods, Layton and her team developed an optimized pipeline for detecting microRNAs in fecal samples and successfully demonstrated its effectiveness in profiling these molecules in mouse stool. Published in Nature Communications, their study offers a reliable approach for isolating and sequencing fecal microRNAs, with potential use as biomarkers for diseases like cancer. Alessio Naccarati, a molecular epidemiologist at the Italian Institute for Genomic Medicine who was not involved in the study, praised the work, noting its promising implications for diagnostic research.
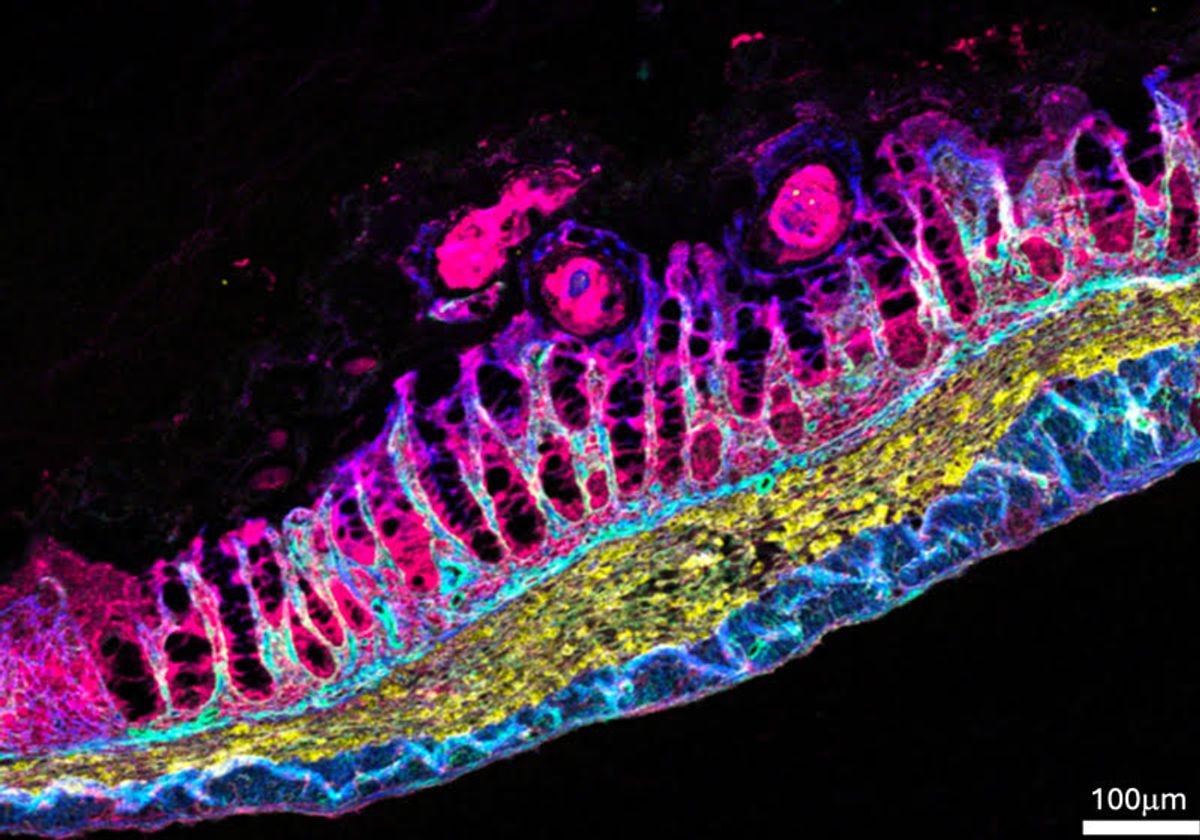
Figure 1. Standardized Fecal microRNA Pipeline Detects Fibrosis Markers in Infected Mouse Intestine
To create a standardized approach for isolating and sequencing fecal microRNAs, Layton and her team tested and refined various protocol components. They determined the optimal storage temperature for fecal samples and fine-tuned PCR settings to build high-quality cDNA libraries. Their optimized method outperformed three commonly used techniques, producing more microRNA reads and detecting a wider variety of microRNAs. With this improved protocol, the team examined microRNA expression in mice infected with the intestinal parasite Trichuris muris. Infected mice showed distinct changes in microRNA levels, including increased miR-200c and decreased miR-29a. Analysis of these microRNAs’ target genes suggested they may be involved in fibrosis, pointing to their potential use as biomarkers for fibrotic diseases like inflammatory bowel disease. Figure 1 shows Standardized Fecal microRNA Pipeline Detects Fibrosis Markers in Infected Mouse Intestine.
To investigate how these microRNAs influence fibrosis, the researchers treated cultured fibroblasts with mimics or inhibitors of the identified microRNAs. Inhibiting miR-29a led to increased collagen production, suggesting that the drop in miR-29a seen after parasitic infection contributes to fibrosis. Similarly, introducing a miR-200c mimic also boosted collagen output, aligning with the idea that elevated miR-200c levels post-infection may promote fibrotic activity.
Finally, Layton and her team validated their findings in vivo by examining intestinal tissue from both healthy and T. muris-infected mice. Infected mice showed significant fibrosis, along with increased levels of fibrosis-related microRNAs—supporting the idea that these small RNAs contribute to infection-induced fibrosis. Layton noted that seeing these results in live tissue confirmed the potential of their technique to identify meaningful disease biomarkers.
“The fact that they were able to link infection-related changes to microRNAs is likely the most significant and novel aspect,” said Alessio Naccarati, a molecular epidemiologist at IIGM who was not involved in the study. He added that further research under various conditions could help assess how effectively this protocol identifies disease biomarkers.
Barbara Pardini, also a molecular epidemiologist at IIGM, praised the clarity of the framework presented in the study but noted that the method could be expensive.
Layton acknowledged these concerns and suggested that future efforts could aim to make the pipeline more high-throughput and cost-effective. “There’s definitely room for further optimization, and I hope this initial work contributes to that progress,” she said.
References:
- https://www.the-scientist.com/navigating-the-complexities-of-cell-culture-72739
Cite this article:
Janani R (2025), Profiling MicroRNA in Feces to Investigate Gut Health and Disease, AnaTechMaz, pp. 406